By Edward Deacon, SciTech Digital Editor
Researchers from Bristol University’s School of Chemistry have shown that an interactive virtual reality (VR) technique is a ‘useful and effective’ tool for developing drugs to fight the SARS-CoV-2 virus.
The novel technique will be useful in finding enzyme inhibitors – molecules that inhibit a certain protein from functioning and being able to reproduce – for an enzyme found in SARS-CoV-2 known as the main protease (Mpro). Such inhibitors would prevent the virus from reproducing, and by doing so, act as effective drugs.
Understanding how a drug molecule binds to a certain protein enables researchers to devise alterations to the drug to make it bind more tightly; this is significant as the tightness with which a drug binds to a target is a key predictor of its effectiveness.
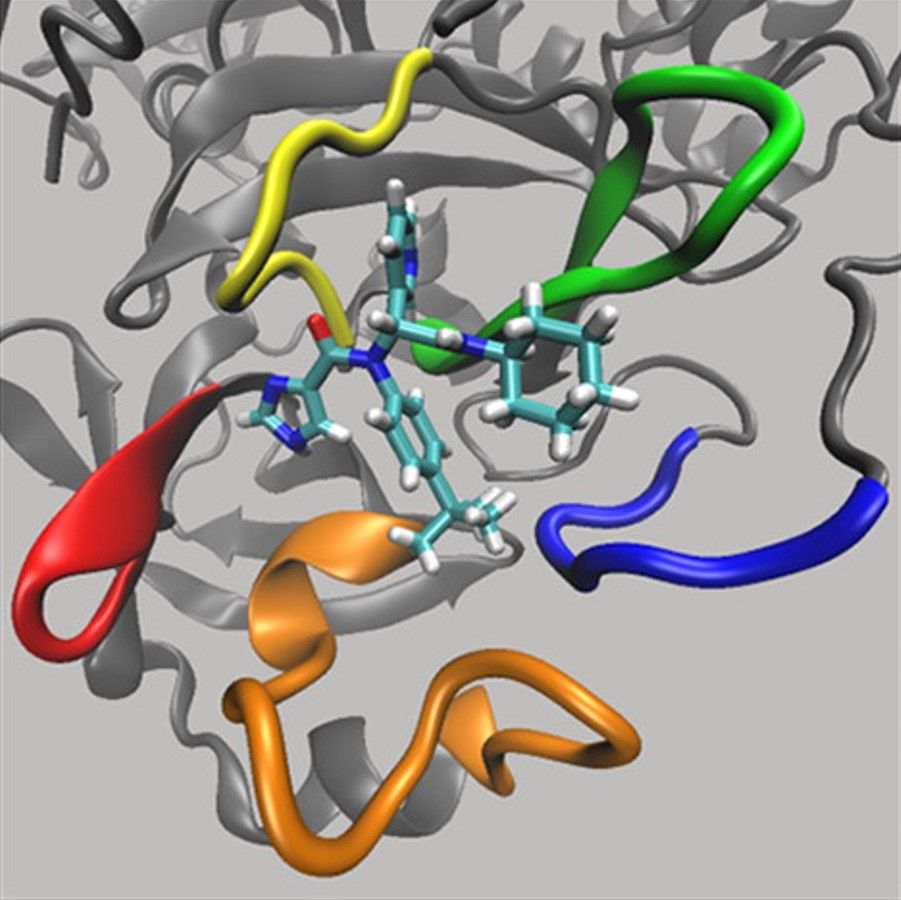
Lead author of the study, Professor Adrian Mulholland, explains that the ‘interactive virtual reality can model how viral proteins and inhibitors bind to the enzyme’, helping researchers to ‘design and test new potential drug leads’.
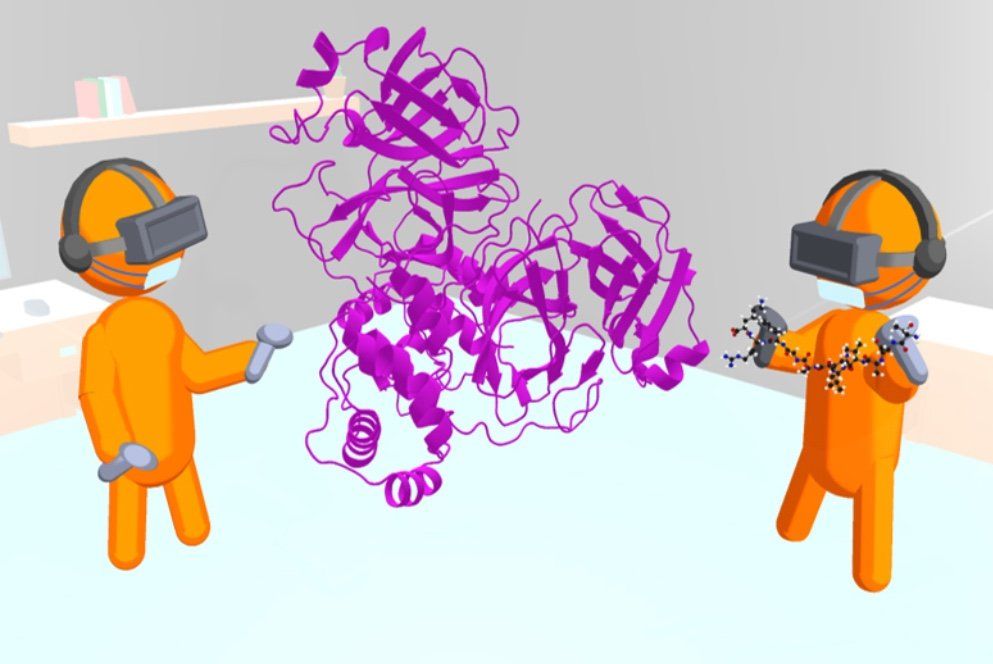
The VR tool for interactive molecular dynamics (iMD) simulations – called Narupa – has been made open source meaning it is freely available for anyone to download. Professor Mulholland noted that: ‘We are sharing these models with the whole community.’
The Bristol researchers created a three-dimensional model of Mpro and used the VR simulations to ‘step inside it’ to visualise the bonding of molecules to the enzyme with the detail of individual atoms.
Our iMD-VR tools will be a valuable resource, enabling virtual collaboration for the international drug discovery community, helping to predict how potential drug leads bind to SARS-CoV-2 targets
Commenting on the study, Professor Mulholland said: ‘There are currently many efforts globally aimed at identifying drug leads for COVID-19. Our iMD-VR tools will be a valuable resource, enabling virtual collaboration for the international drug discovery community, helping to predict how potential drug leads bind to SARS-CoV-2 targets.
An exciting aspect is that it also allows researchers to collaborate in new ways: using cloud computing, they can tackle a drug discovery problem together at the same time when in they are in different locations – potentially even in different countries – working simultaneously in the same virtual molecular environment.’
University of Bristol COVID-19 research updates
Trump’s tweets led to ‘systematic diversion’ away from negative publicity, Bristol study finds
The cloud computing for the online framework of Narupa was hosted by Oracle for Research. Vice President, Alison Derbenwick Miller, said: ‘We are delighted that Oracle's high-performance cloud infrastructure supported the development of this innovative framework’.
And commending the Bristol team, she said: ‘Computational modelling of how drugs bind to the SARS-CoV-2 spike protein has been critical in advancing the global fight against the pandemic. Narupa takes that modelling to an entirely new level with molecular dynamics simulations in virtual reality.’
Featured Image: University of Bristol
Are you impressed by this new drug discovery technique?